Trendmain - Obtain Principal Components from a series of 2D Measurements
- Launch using
trendmaingui.exeortrendmaingui.app. In the left side bar, there are four modes to choose from:basic,all_purpose,nmr, andmovie. They function similarly with minor differences.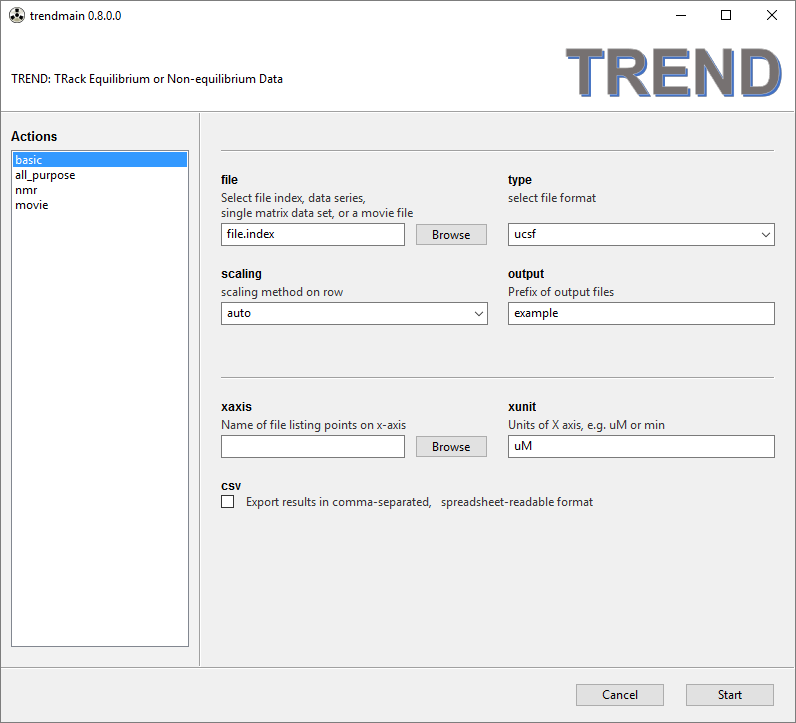
- The
basicmenu can be used for simple PCA work. A series of 2D data files (or an index file listing the series of data files) can be read by thefileFile Chooser widget. Note thefileFile Chooser can do two things: If input file names contain numbers, TREND can extract numbers from the file names and sort these names numerically. A corresponding file index (file.index) will be generated automatically. Or you can make a file index manually with a text editor according to the format described in the manual. - Choice of file format, scaling methods (on rows), prefix of output file names can
also be specified. The lower half of the panel shows optional arguments
(which can be left blank). A file list the tick marks for the X-axis can be
selected by the
xaixsFile Chooser. The units of the X-axis can also be specified and will take effect if a file listing the points of the X-axis measured is selected inxaxis. When thecsvcheckbox is turned on, the PCA results will be exported in comma-separated (CSV) format that can be read by spreadsheets such as Excel or OpenOffice. Thereportcheckbox is on by default. This generates an HTML report of arguments used by main script, links to files generated by PCA (PC values, normalized PC1, other files for plots, the S matrix, etc), and illustrations of the scree plot, normalized PC1, first 3 PCs, and autocorrelation coefficents.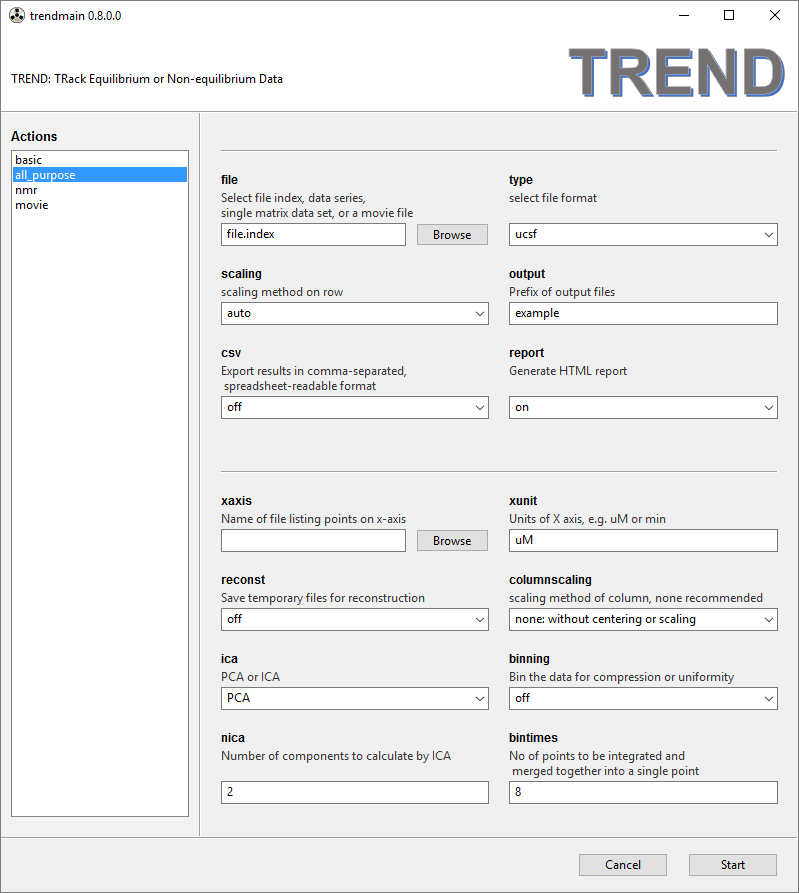
- The
all_purposemenu provides additional options. When thereconstoption is turned on, temporary files are saved for later use in reconstruction of the data from the components specified. Scaling of columns can be specified, but should not be necessary if the rows are scaled. If the input file type is Fourier-Transformed NMR spectrum (NMRPipeft2format or SparkyUCSFformat), the spectra can be "binned" for compression or uniformity. Thebintimesparameter for this can be speicified. Detail descriptions can be found in the manual. In theall_purposemenu, ICA (independent component analysis) can be selected instead of PCA. WhenICAis chosen, the number of independent components to be calculated should be specified ( Jia Xu and Steven R. Van Doren, Tracking Equilibrium and Nonequilibrium Shifts in Data with TREND. Biophys. J. 2017, 112,224-233
The all_purpose menu supports various formats, including JCAMP-DX, which
is a general format for exchanging and archiving data from many instruments,
inlcuding but not limited ot IR, Raman, Uv-Vis, Fluroescence, NMR, EMR,
mass spectrometery, and chromatography.
- The
nmrmenu is for analysis of NMR spectra. It is similar to theall_purposemenu but offers selection of data only in these formats: JCAMP-DX, NMRPipe fid and ft2, Sparky ucsf and Sparky peak list as sparkylist, Bruker Topspin FIDs and spectra as brukerfid and brukerspectra, and Agilent (Varian) VnmrJ FIDs and spectra as agilentfid and agilentspectra. The Topspin and VnmrJ formats are saved as directories, in contrasting them with the other formats that save data as individual files. Therefore adirDirectory Chooser widget is provided for the Topspin and VnmrJ data directories. Its usage is similar to thefileFile Chooser widget described previously. Note space is NOT allowed in the Topspin or VnmrJ directory names. A manually made directory index, which is in the same form offile.indexcan still be read by thefileFile Chooser for parsing Topspin and VnmrJ directories. See manual for details.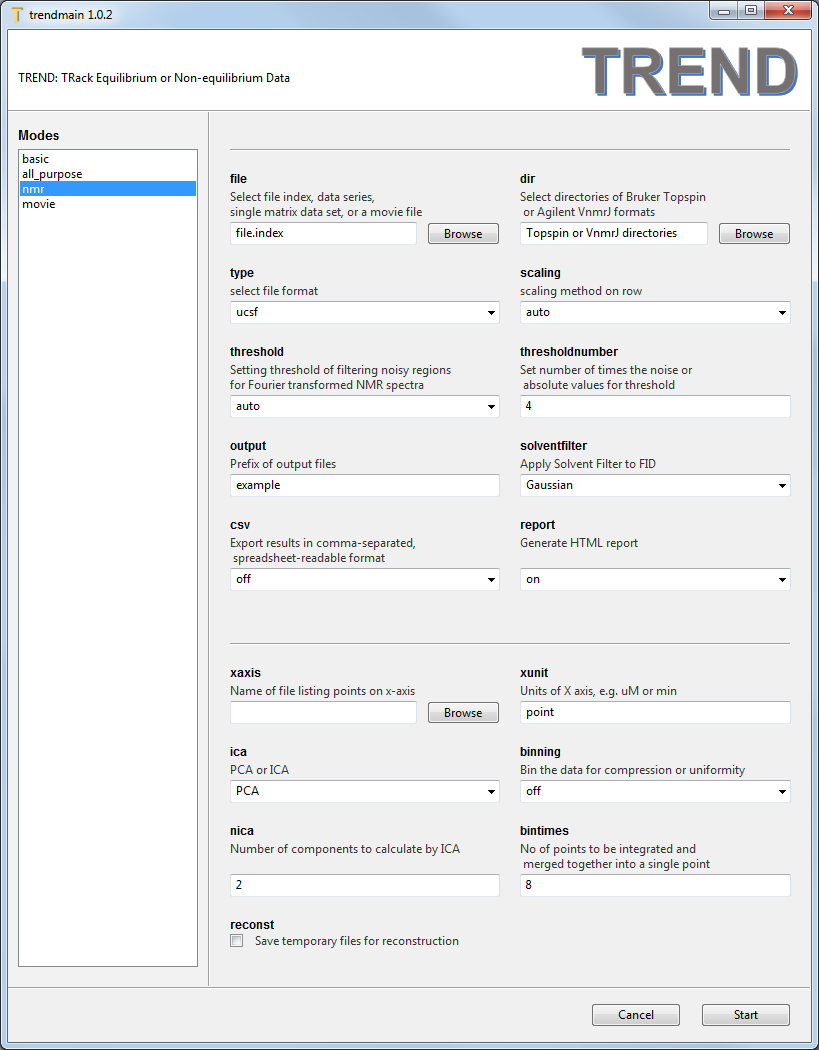
The default scaling method of rows of the data matrix is auto, which is recommended for series of NMR spectra in fast or slow exchange regimes. pareto is recommended for spectra with intermdiate exchange. noscaling should be acceptable for all exchange regimes. Details of choosing scaling method and setting the threshold to filter out noise are given in the manual and Jia Xu and Steven R. Van Doren, Binding Isotherms and Time Courses Readily from Magnetic Resonance. Anal. Chem. 2016, 88 (16), pp 8172-8178thresholdandthresholdnumbercan be set for filtering noise out of NMR spectra. The three ways to set thethresholdare auto, absolute, and the number of times the noise level. In auto mode, the program determines noise from the first spectrum and sets the threshold as 4-fold the noise level forautoscalingand 0.5 times the noise level forParetoscaling.solventfilterprovides options of applying solvent filter to FID data, none, Gaussian, sine-bell, sine-bell-square are provided. rectangular filter is provided in the command-line mode. See the manual for definitions of solvent filter.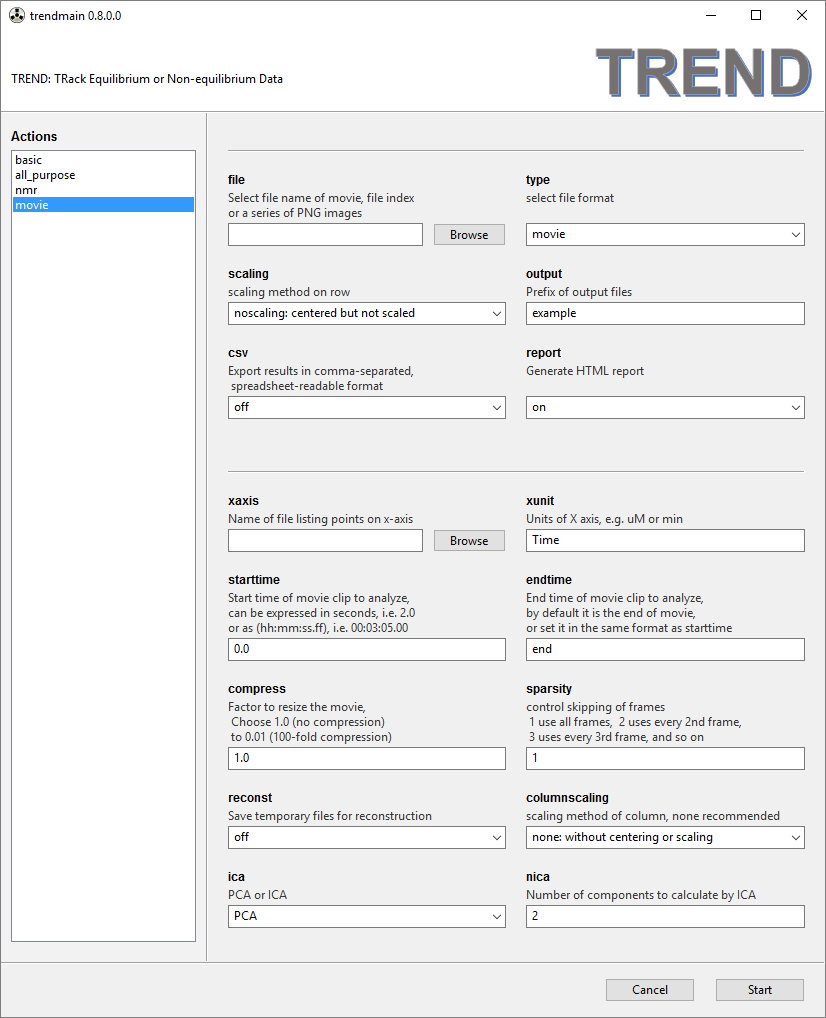
- The
moviemenu is for processing movies or time-dependent PNG image series. The recommended scaling method of rows is noscaling. A file listing tick marks for the x-axis can be specified. When processing a movie, time will be automatically extracted from the movie and saved asmovie_time.txtfor use as x-axis. The movie (but not series of PNG images) can be resized to make it smaller by skipping frames of the movie and setting the parameterscompressandsparsityaccording to the manual. TREND can select a certain portion of a movie by settingstarttimeandendtimeparameters. The numeric format for setting the start and end time can either be floating point numbers of seconds (e.g.0.2), orhh:mm:ss.ff, such as00:03:05.00. The default values for thestarttimeandendtimeare0.0andend, which do not trim the movie.