Main Script - Obtain Principal Components from a series of 2D Measurements
This describes the options of the CLI (Command Line Interface) program (which underline the GUI version described above)
GUI:
trendmaingui.exein Windows and Linux,trendmaingui.appin OS XCLI:
trendmain.exeon all platformsSynopsis
trendmain.exe -t [ucsf/fid/ft2/png/movie/txt/sparky/csv/excel/…] -f [file index] -r [threshold] -o [output prefix name] -s [scaling methods] [-x [xaxis file]] [-u [Unit]] [--report]Description
trendmain.exereads a series of NMR spectra, FIDs, png images, the Joint Committee on Atomic and Molecular Physical data - Data eXchange (JCAMP-DX) format, plain text files, comma-separated (CSV) or Excel spreadsheets listed in the index file.trendmain.exealso reads and process a single file, including movie, an Excel spreadsheet, a comma-separated (CSV) text file, a JCAMP-DX file with multiple blocks, or a plain text file. TREND preprocesses them, applies singular value decomposition to provide U, s, and V matrices (Jia Xu and Steven R. Van Doren, Binding Isotherms and Time Courses Readily from Magnetic Resonance. Anal. Chem. 2016, 88 (16), pp 8172-8178). Its routine mode is to provide principal components (PCs) from the first few rows of the V matrix. TREND also has options to perform PCA reconstructions of the original data series or to perform independent components analysis (ICA). The S and V matrices and normalized PC1 values are output as text files in the names ofprefix-s.txt,prefix-vt.txt, andprefix-pc1.txt, which reports the binding isotherm. Additional PCs can be requested.Typical usage with NMR spectra:
trendmain.exe -t ucsf -f file.index -r auto -o outfile -s auto --reportOptions
-t [ucsf/fid/ft2/png/movie/txt/complextxt/csv/complexcsv/singlematrix]-tspecifies the input file format, the default format is .ucsf for NMR spectra. The options for input file formats are:NMR file options
- ucsf Sparky ucsf format (default)
- fid NMRPipe fid format
- ft2 NMRPipe ft2 format
- png image format (needs Scipy module)
- brukerfid Bruker Topspin FID format (
fidorserfile in the experimental directory) - brukerspectra Bruker Topspin processed spectra (
1r(1D) or2rr(2D) in the/pdata/1subdirectory in the experimental directory . Note currently the processed spectra must be saved by setting processing number as 1 in the Topspin experimental directory - agilentfid Agilent VnmrJ (OpenVnmrJ) fid. (
fidfile in the data directory) - agilentspectra Agilent (Varian) VnmrJ (OpenVnmrj) fid. (
phasefilein thedatdirsubdirectory. Note in order to makephasefilereadable by third-party software including TREND, set trace='f1', display the full spectrum, and use the VnmrJflushcommand are required. Otherwise thephasefilewill be all zero values. See the Sparky manual for details. Note TREND supports on-the-fly analysis on Bruker and Agilent spectrometers. Please note that TREND identification of principal comopnents can be disrupted by a change in acquisition parameters or an outlier among the spectra. Consequently, all spectra or data frames to be analyzed by TREND should be collected and processed under identical conditions. Therefore, in the case of NMR spectra, we highly recommend using the same set ofNMRPipescripts to convert Bruker or Agilent data into NMRPipe fid and ft2 formats.
Other input formats: text, spreadsheet, JCAMP-DX spectra, and movies
- txt Data stored in plain text format delimited by spaces, this option could be used for other spectroscopy methods such as time- dependent 2D-IR.
- csv csv format is supported by Excel, OpenOffice and many software for spectroscopy instruments.
- complextxt Similar to plain text format (
txt) but using complex number in the form of0.00+0.00j. - complexcsv Similar to csv format. However, each cell should be in
the format of
0.00+0.00j. - excel Microsoft Excel format (xlsx is preferred, the older xls
format is also accepted). All cells must be float or complex numbers.
The complex number should be in the form of
0.00+0.00j. By default an Excel file contains three sheets, only the first sheet is analyzed. Excel with multi-sheets can be processed assingleexceloption. - sparky or sparkylist Sparky list format. The 1st column lists any residue assignments. The 2nd and 3rd columns are F1 (often 15N) and F2 (often 1H) chemical shifts, respectively.
- singlematrix This mode serves as a general PCA tool to process a single matrix in the text format delimited with spaces.
- singlecsv Similar to
singlematrixformat but incsvformat - singleexcel Similar to
singlematrixandsinglecsvformats but in thexlsxorxlsformats. Besides, a series of 1D or 2D dataset can be stored in a single Excel file containing different, sequentially-ordered sheets. - PythonNPY is a standard binary file format NPY for Python Numpy.
- Matlab MAT-Files. However the Matlab file should contain just 1 variable. See save command of Matlab.
- movie Common video formats, such as
.ogv,.mp4,.mpeg,.avi,.mov,.webm - jcamp JCAMP-DX is a general format for exchanging and archiving
data from many instruments, including but not limited to IR, Raman,
Uv-Vis, Fluorescence, NMR, and EMR. The data stored in JCAMP-DX
files can be spectral plots, contours, or peak tables. JCAMP-DX
is very flexible in order to support most kinds of computerized
instruments. As a result, it is impractical to support all
existing JCAMP-DX variants. However, TREND supports most common
JCAMP-DX formats. The digital data in JCAMP-DX can be AFFN
(ASCII FREE FORMAT NUMERIC) form or ASDF (ASCII SQUEEZED
DIFFERENCE FORM). TREND supports decoding compressed data,
including
PAC,SQZ,DIF,SQZDUP, andDIFDUP. Two most common tabular data form,(X++(Y..Y))and(XY..XY)are supported. TREND reads a series of JCAMP-DX files, or a single JCAMP-DX file with one or multiple blocks. TREND supportsNTUPLEformat (introduced by JCAMP-DX 5.0 standard), which is designed for multi-dimensional techniques with data sets of multiple variable. For example, JCAMP-DX NMR usesNUTPLEto show mixed real/imaginary FID data sets. See http://www.jcamp-dx.org/, https://badc.nerc.ac.uk/help/formats/jcamp_dx/ and http://wwwchem.uwimona.edu.jm:1104/spectra/testdata/index.html for details of JCAMP-DX formats.
Reading a collection of files or spectra:
-f [fileindex/filename]-freads file index and is required[fileindex]is a text format index referring to the series of NMR spectra, images, lists, or directories (for Bruker Topspin and Agilent VnmrJ data). The default file index name isfile.index. An example is a series of five .ucsf files named numerically. The index file can be named asindex.ucsfand includes the following lines in the example:
Another example is a series of Bruker Topspin directories on a Windows machine. Absolute paths are supported, as well as relative path, which is shown in the example ofUCSFfiles. This "directory list" can be used for both brukerfid and brukerspectra. For brukerfid data, TREND will readfidorserdata from the listed directories. For brukerspectra data, TREND will read1ror2rrfiles from thepdata/1subdirectorires. Therefore, the number of the pdata subdirectory for processed files must always be set as1.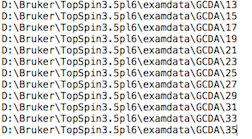
file.indexfor Agilent VnmrJ format could be used in a simmilar manner, except its spectra are saved in thedatdirsubdirectories.- [filename] can be the name of a singlematrix data set or a movie file for processing.
Preprocessing options:
-s [none/noscaling/auto/pareto/]-sspecifies scaling method applied on rows of the data matrix.- none means skip centering and scaling of the rows of the data matrix.
- noscaling means do data centering without scaling. It should be accetable in most conditions.
- auto means Autoscaling. It treats all peaks or features as equally important but inflates the measurement errors. It is recommended for NMR spectra in fast and slow exchange regimes.
- pareto means Pareto scaling. It reduces the importance of large peaks but enhances the low, broader peaks. It is recommended for NMR spectra in intermediate exchange.
- Definitions of the scaling methods are given in: J. BMC Genomics 2006, 7, 142. Other scaling options mentioned in this paper, such as vast scaling, range scaling, level scaling are also provided.
-r [auto/1e5/3T/5t]-rsets the threshold for filtering low intensity regions. The threshold is meaningful when processing 2D NMR spectra (ucsf, ft2), but not for FIDs or images in png format. There are three ways to set the threshold: auto, absolute, and manual (set the number of times the noise level).- auto In auto mode the program determines noise from the first spectrum and set threshold as 4 times of the noise level for autoscaling and 0.5 times the noise level for Pareto scaling.
- 1e5: When the arguments are set as integral numbers or floating point
numbers , such as
1e5or100000. This is used to specify the threshold value using the numbering scale of the measurement. - 3T or 5t: Numbers with suffix
Tortannotates times of noise level of the first spectrum. In this mode threshold is set as 3 or 5 times of noise level of the first spectrum. Three to seven-fold (3T to 7T) is the recommended range for NMR spectra.
-o [output prefix name]-ospecifies the prefix of output filesprefix-s.txt,prefix-vt.txt,prefix-pc.txt, andprefix-PC1.txtfrom PCA.prefix-pc.txtreports all principle components as successive vertical columns.prefix-PC1.txtreports the first principal component as a single vertical column, which in a titration is the binding isotherm.For example, the option
-o testgenerates output files named astest-s.txt,test-vt.txt,test-PC.txt,test-PC1.txtprefix-s.txtandprefix-vt.txtsave S and VT matrices in the SVD calculation. U matrix is by default omitted because its usually big size.When ICA is used instead of PCA (
-ioption is on), the prefix includesprefix-vt.txt, which is unmixing matrix, andprefix-IC.txt, which reports normalized independent components as successive vertical columns.--csvWhen--csvis turned on, data inprefix-s.txt,prefix-pc.txt,prefix-PC1.txt, orprefix-IC.txtare generated as CSV files, which can be read by software such as Excel or OpenOffice. The corresponding file names areprefix-s.csv,prefix-pc.csv,prefix-PC1.csv,prefix-IC.csv. In csv files (exceptprefix-s.csv), data is organized as sucessive vertical columns, where the first column represents component number. A table header is added, such as# Component, PC1, PC2, PC3 ....-b
Traditional uniform binning is applied for NMR data when-boption is turned on, it does not improve accuracy but may be useful when the input dataset is too large. By default-boption isoff.bintimesoption needs to be set in this condition.--bintimes [binning time]
Number of points to be integrated and merged together into a single point. The default is 8. For example, if the size of a 2D spectra is 1024*2048, setting--bintimes 8causes size of binned spectra to become 1024/8 2048/8 = 128 256--columnscaling [none/noscaling/auto/pareto]--columnscalingspecifies the scaling method applied on columns of the data matrix. Options and meanings of scaling methods are defined in the-soption. By default--columnscalingis set as [none].--solventfilter [none/Gaussian/sine-bell/sine-bell-square]
--solventfilterapplies solvent filter to FID signals infid,brukerfid,agilentfidandjcamp-dxformat
none means no water filter will be applied
Gaussian applies solvent filter with Gaussian filter
sine-bell applies solvent filter with sine-bell shaped filter
sine-bell-square applies solvent filter with squared sine-bell shaped filter
rectangular applies solvent filter with boxcar filter
See nmrglue reference guide and Marion et al, J Mag Reson 1989 84, 425-430 for details.-ior--ica
This option uses the independent component analysis (ICA) module instead of PCA.--nica
When ICA is used, the number (n) of components to be calculated must be set as--nica n. The default n is 2.--reconst--reconstis an option for reconstruction. It has no effect on the calculation, but dumps necessary files for reconstruction in the future--keepfilteredis an option for 2D NMR spectra reconstruction when--reconstis on. When the input files are 2D NMR data such as ucsf or ft2, a threshold can be set using the-roption. All data points below the threshold are filtered out.--keepfiltereddumps the filtered data points to a file for reconstruction purpose. This file is not required because reconstruction of PCA can be done without the filtered data points.-x [xaxisfile]-xspecifies the name of file containing the list of tick marks to be used for the x-axis. It is optional. By default this option is off and x-axis ticks are integers. Whenxaxisfileis specified, numbers within it will form the x-axis. The number of rows inxaxisfileshould identical toindexfile specified by the-f [fileindex]option oftrendmain.exe
For example,concentration.txtlists the five concentrations of a titration, one per line: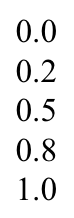
-u [Unit]-uspecifies label placed on the X-axis for plotting. [Unit] can be any string.--report--reportgenerates an HTML file named asprefix-report.htmlthat reports the results. The report lists arguments used for calculation and files created. It plots the first 3 PCs or ICs without normalization. It also plots scree plot as well as autocorrelation coefficences that represent the smoothness of component curves. For more control of plotting of principal or independent components display, please usetrendplot.exe.Movie processing options:
--compress [compress factor]
When processing movie the video can be resized by [compress factor] to reduce computational cost. For example, 0.8 means the video size will be resized to 80%. [compress factor] can be cho sen from the interval (0, 1.0]. By default [compress factor] is set as 1.0, which means use all the data, i.e. don't compress the data.--sparsity [sparsity factor]
This option controls skipping frames of the input video file by picking up every n-th frame from the video, where n is specified by [sparsity factor]. For example, a [sparsity factor] of 1 means all frames will be used. 2 uses every second frame, 3 uses every third frame, and so on.--starttime [start time]
TREND supports making a subclip of the input movie by setting its starting and ending time. The default value for--starttimeis 0.0. The numerical format for setting the start time and end time could either be floating point numbers of seconds (e.g.0.2stands for 0.2 s), orhh:mm:ss.ff, such as00:03:05.00, which means 3 minutes and 5 seconds.--endtime [end time]
This option sets the ending time for the subclip of input movie. By default it is set asend. When--starttimeand--endtimeare set as the default (0.0andend, respectively), the whole input video will be analyzed. Otherwise a subclip video will be analyzed and exported asfrom-starttime-to-endtime_movieclip.mp4, orfrom-starttime_movieclip.mp4if the--endtimeis set as the defaultend.