Simple Example: Determine binding isotherm from NMR spectra in UCSF format
GUI USAGE
- Choose
nmrtab oftrendmainguiand press browse button highlighted by the box.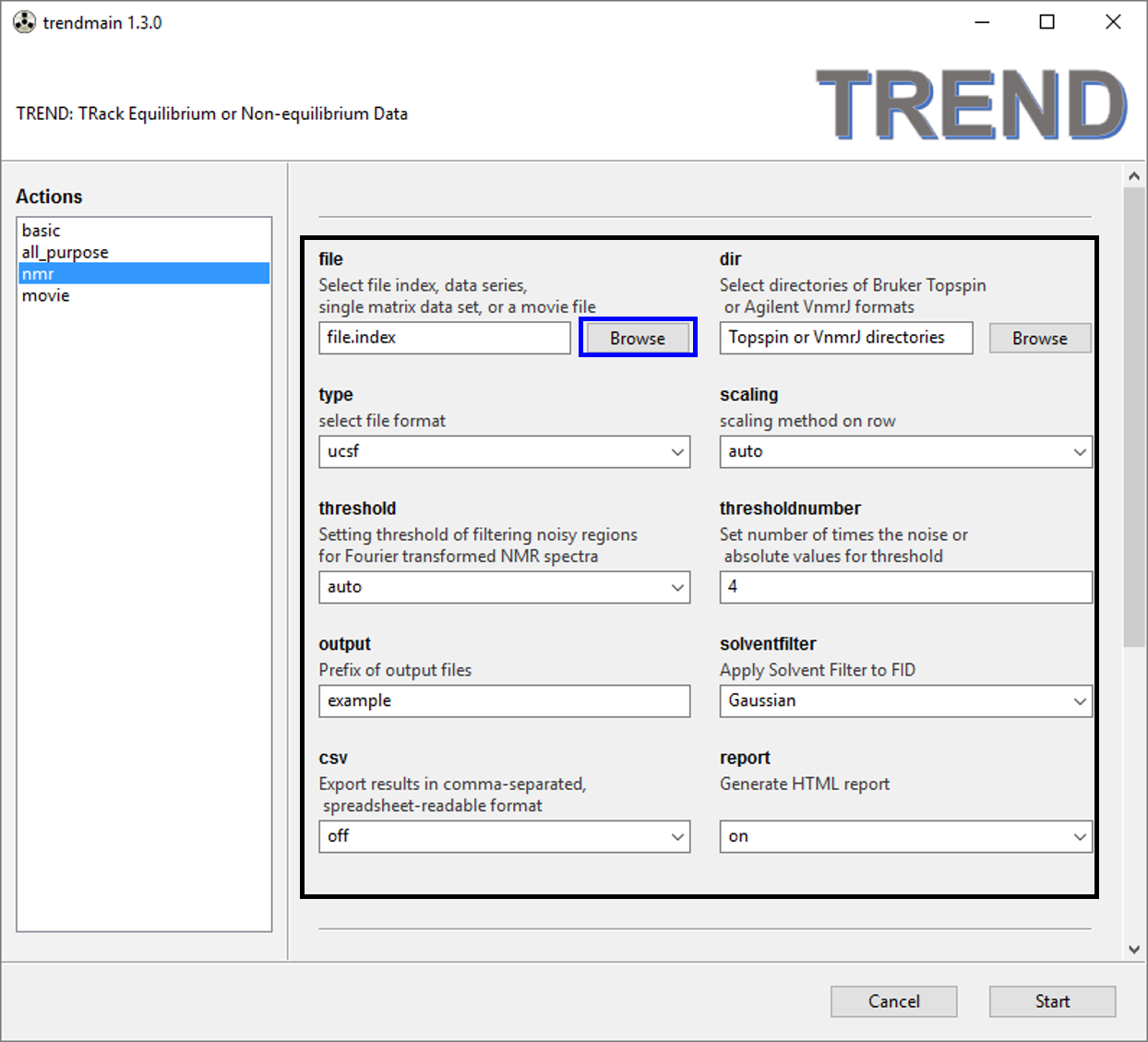
Choose a series of Sparky format spectra (.UCSF files) and TREND will sort them numerically.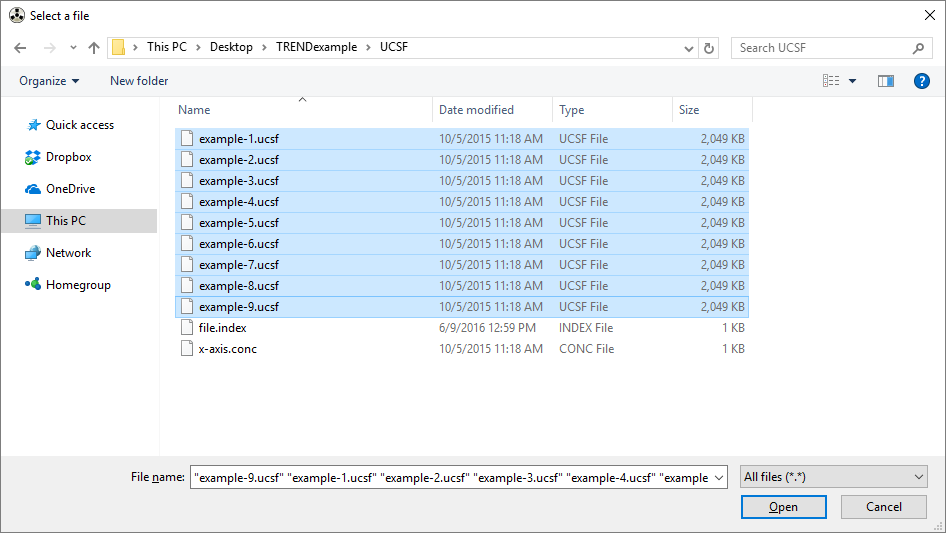
- Specify required arguments including file format, method of scaling rows, the prefix of output files, threshold for smallest signals to be retained (x-fold the noise), whether to export CSV-format spreadsheet files, and whether to generate an HTML report. The default settings are recommended.
- Optional arguments are highlighted within the box with dashed line.
See the manual for details. Here we specify
xaxisandxunits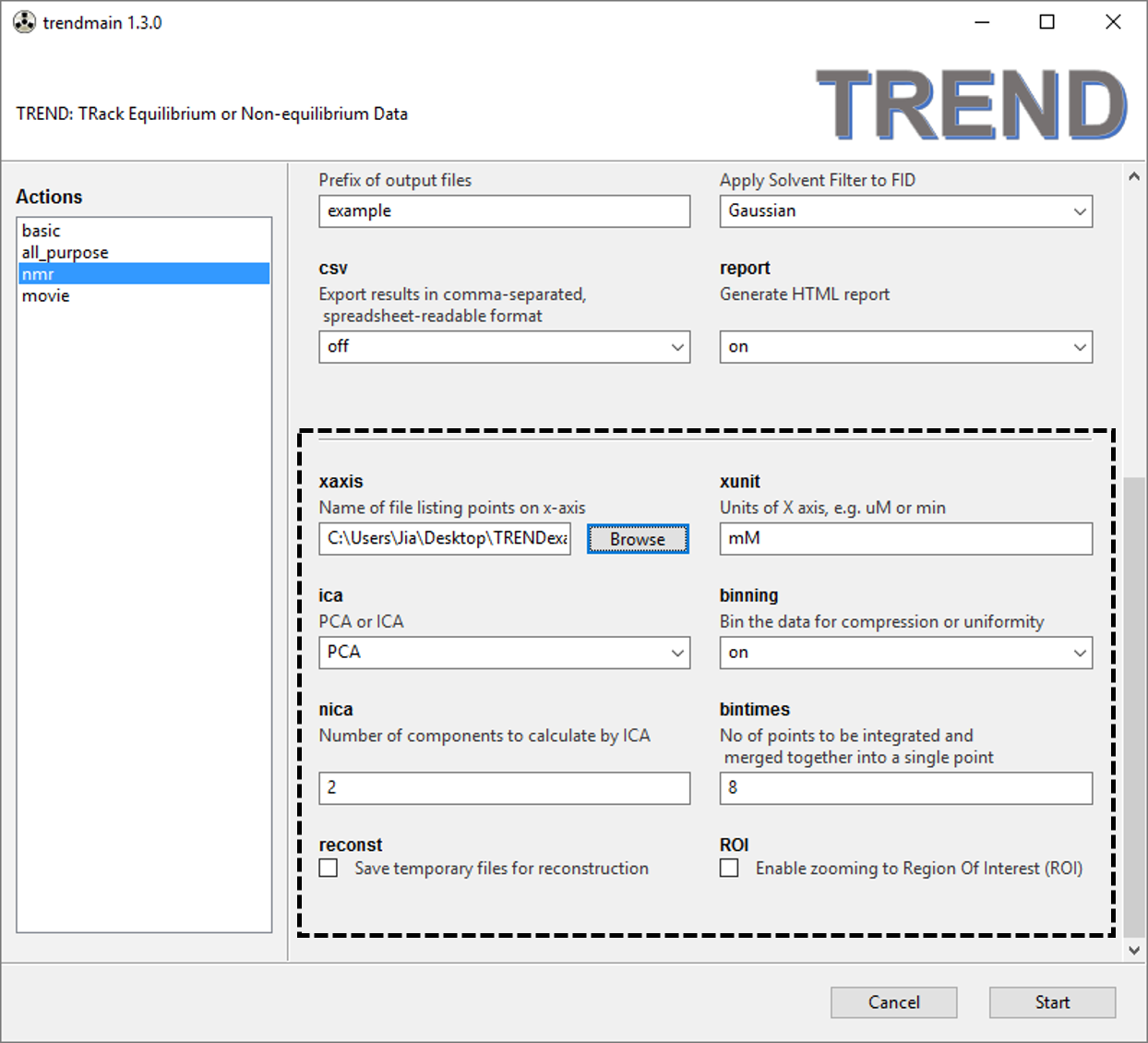
- Press start button, TREND will run for a while and finish.

After
trendmainfinishes, an HTML report is generated. It includes the arguments just used, scree plots, and plots of principal components, etc. Please see the manual for details.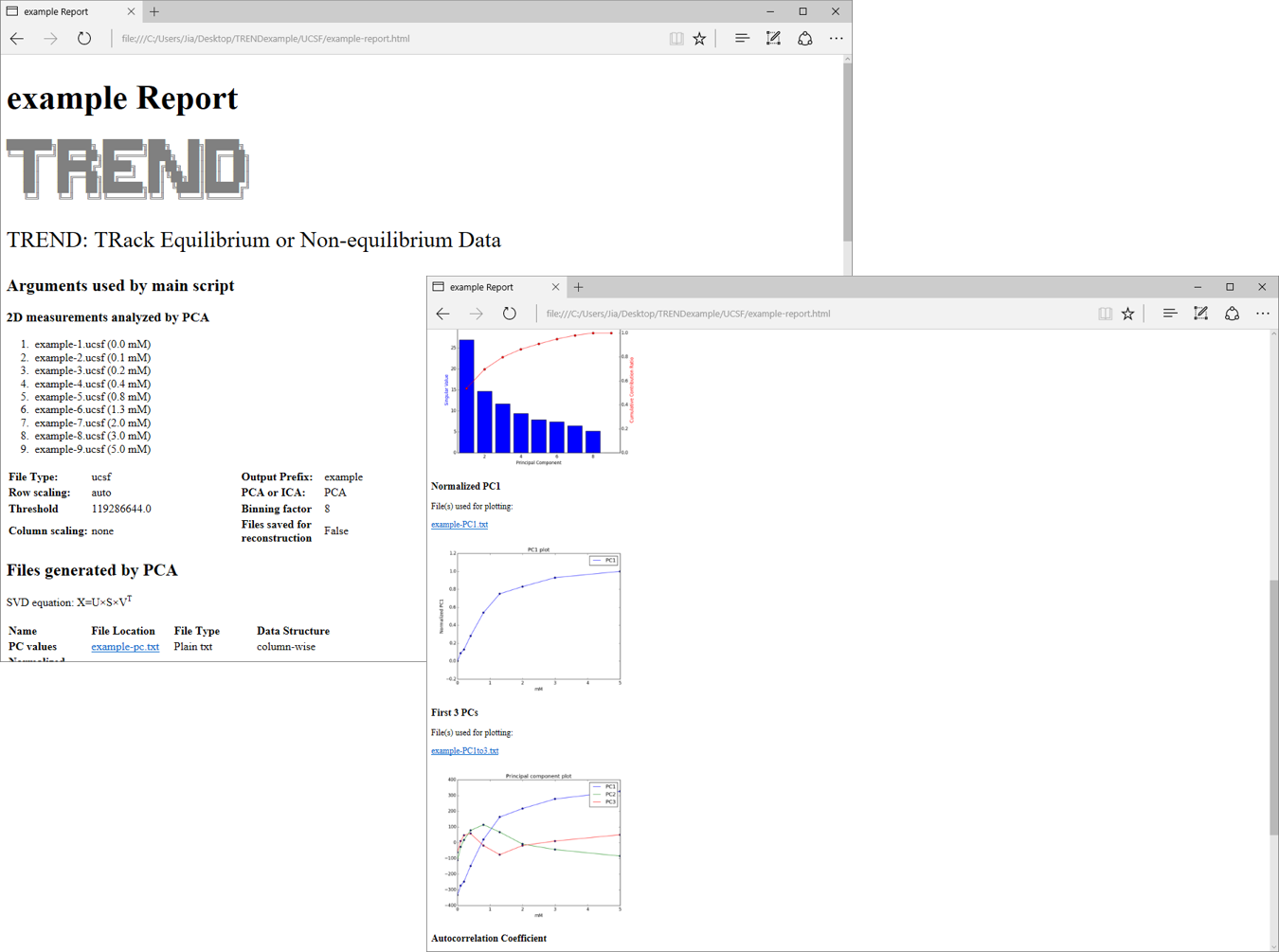
Corresponding shell command
trendmain.exe -f file.index -t ucsf -s auto -o example -r auto --reportCitation
If you use TREND in your research, please cite these references in resulting publications:
Jia Xu and Steven R. Van Doren, Binding Isotherms and Time Courses Read ily from Magnetic Resonance. Anal. Chem. 2016, 88 (16), pp 8172-8178
and
Jia Xu and Steven R. Van Doren, Tracking Equilibrium and Nonequilibrium Shifts in Data with TREND. Biophys. J. 2017, 112,224-233